Hypoxia, the formation of reactive oxygen species (ROS) and subsequent oxidative stress in synovial tissues are key events in the pathogenesis of RA1. We provide evidence here that CBP and p300 are critical regulators of the adaptive response of SF that integrate the transcriptional and functional regulation of stress response pathways throughout the cell.
CBP and p300 are HATs and H3K27ac mark writers, which activate post-translational histone modifications present in enhancers and promoters20. Among the CBP/p300 target proteins are several non-histone proteins in addition to histones, including many transcription factors and signaling effectors16.21. Despite their high protein sequence homology, CBP and p300 have distinct individual functions identified by our own and other studies12,13,14.
In SF, persistent H3K27ac in inflammatory gene promoters was associated with long-lasting and persistent expression of the corresponding genes22. We recently demonstrated that p300 is the major HAT in SF that exerts a pro- and anti-inflammatory role. This is in contrast to CBP, which exerted anti-inflammatory effects upon silencing, and specifically regulated TNF-induced interferon signature gene expression14. Although we have ruled out that silencing p300 additionally reduced CBP and vice versa, we cannot completely rule out other potential off-target effects after our silencing approach. Krosel etc already. have shown that inhibitors targeting the HAT or bromodomain of CBP/p300 closely resemble the effects of p300 silencing, including increased expression of TNF-induced proinflammatory gene expression14.
Our RNAseq data provide evidence that the number of p300-regulated target genes is greater than that of CBP-regulated target genes, also in terms of stress response. While the cellular response to oxidative stress and autophagy was co-regulated by CBP and p300, genes associated with response to oxygen levels, hypoxia, and pathways associated with proteasome regulation and function were specifically enriched upon knockdown of p300. In pathways co-regulated by CBP and p300, we identified several genes that were regulated in opposite directions. These results point to individual functions of the two enzymes at the level of target genes, similar to what we have already observed for many inflammatory genes14. Furthermore, a small number of measured CBP and p300 target genes, namely BCL2, SOD3 and HDAC6, could be regulated in a joint-specific manner, as indicated by our Real-time PCR results in a limited number of samples for each joint site. Frank-Bertoncelj et al. have previously shown that H3K27ac is one of the mechanisms controlling the joint-specific expression of homeobox (HOX) transcription factors in SF from different locations7. To draw a definitive conclusion whether stress-associated target genes are regulated in a joint-specific manner, larger numbers of SF from different joints would be needed, together with H3K27ac ChIPseq data in unstimulated and TNF-stimulated SF from different joints.
In addition to the differential roles of CBP and p300 in regulating target gene expression, we showed here a differential regulation of CBP and p300 by stimulating SF with 4-HNE and TNF (Fig. 6). These factors are present in the synovial microenvironment in RA and mimic the oxidative stress and inflammation, respectively. While 4-HNE and TNF, similar to H2O2suppressed the expression of p300, CBP was not affected. 4-HNE is a lipid peroxidation product generated at elevated levels of ROS. Levels of 4-HNE are elevated in serum, synovial fluids, and synovial tissues of RA patients, and serum levels of 4-HNE correlate with structural damage such as erosions in the early stage of RA23,24. As mimicked by our silencing approach, the TNF- and 4-HNE-mediated suppression of p300 expression in the synovial RA microenvironment has fundamental consequences for SF behavior. Our datasets from the previous one14 and the present study indicates that reduced expression of p300 was associated with increased expression of many inflammatory cytokines, chemokines matrix metalloproteinases and stress response genes in SF. Among these genes were HK2, a marker indicating the metabolic switch from SF to glycolysis, and VEGF, a pro-angiogenic factor secreted to overcome hypoxia.1.
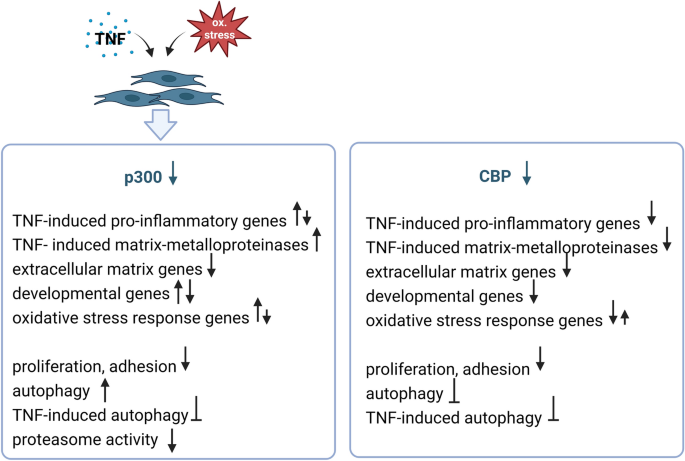
Summary of CBP- and p300-regulated pathways in SF. The expression of p300 but not CBP is down-regulated in synovial fibroblasts after exposure to TNF and oxidative stress. The effects of p300 and CBP silencing are demonstrated based on findings from this and a previous study14. Downward arrows indicate suppressed expression or function, upward arrows indicate increased expression or function. The figure was created by BioRender.com.
TNF stimulation of SF markers of endoplasmic reticulum (ER) was shown to induce stress and autophagy25. Our data suggest that CBP and p300 regulate autophagy at the transcriptional level and influence autophagic flux. The assessment of autophagy in the presence of the lysosomal inhibitor bafilomycin A1 indicated that CBP and p300 regulate autophagy function at different stages within the autophagic process. CBP silencing affected autophagosome synthesis. In contrast, knockdown of p300 induced autophagy in unstimulated SF, and induced a late-stage block of autophagy in TNF-stimulated SF, a condition in which polyubiquitinated proteins in SF accumulated. Accordingly, knocking down p300 only increased cell death in the presence of TNF, as previously indicated14. Kato etc already. have previously shown that autophagy induction partially compensated for reduced clearance of polyubiquitinated proteins in SF after blocking proteasome function, indicating a protective effect of autophagy induction in SF under such conditions5. Here we observed a similar compensatory mechanism after p300 knockdown, which was associated with a suppression of proteasome enzymatic activities and an induction of autophagy. This finding is consistent with a previous study in HeLa cells in which p300 knockdown was associated with reduced acetylation of autophagy-related proteins and increased levels of autophagy.26.
Acetylation and deacetylation of components of the autophagy machinery control all steps of this catabolic process, from autophagosome initiation to LC3 conjugation, cargo assembly, and autophagosome-lysosome fusion27,28. Several classes of acetyltransferases, including CBP and p300, and deacetylases, including sirtuin1, HDAC4 and HDAC6 are involved in the regulation of autophagy27.29. Furthermore, the function of autophagy-related transcription factors, such as transcription factor EB (TFEB), Foxo1 and Foxo3, is regulated by deacetylation28.30. Recently, the increased translation of FOXO3 mRNA has been described to facilitate the initiation of autophagy31. We showed here that in unstimulated SF, FOXO3 mRNA increased after p300 knockdown, a condition in which autophagic flux increased. HDAC6 binds to polyubiquitinated proteins in SF29and promotes autophagy by facilitating autophagosome-lysosome fusion27. On the other hand, HDAC6 was also shown to suppress autophagy by deacetylating TFEB and Foxo130. This could explain the inverse regulation of HDAC6 and ATG5 and ATG16L1 in SF.
A global analysis of the CBP/p300-dependent acetylome in mouse embryonic fibroblasts (MEF) suggested that proteasome functions might also be regulated by these enzymes.21. The majority of proteasome machinery components showed numerous CBP/p300-dependent acetylation sites in regulatory and enzymatic subunits in MEF (http://p300db.choudharylab.org). Furthermore, ATG5 and ATG16L1, two proteins essential for autophagosome assembly, showed CBP/p300-dependent acetylation sites21. Whether proteasome and autophagy components are acetylated in a CBP- and p300-dependent manner in SF remains to be functionally evaluated21. Because CBP/p300-dependent acetylation sites in MEF were analyzed after a combinatorial knockout of both enzymes, it is not clear which of them is the key enzyme in regulating the post-translational acetylation of proteins involved in the regulation of autophagy and proteasome activities.
In summary, we have identified CBP and p300 in particular as critical regulators of stress response pathways in SF, with overlapping and distinct functions within specific pathways. The downregulation of p300 by TNF and oxidative stress provides a mechanism underlying SF activation in the synovial microenvironment.
Leave a Reply